Selected Projects
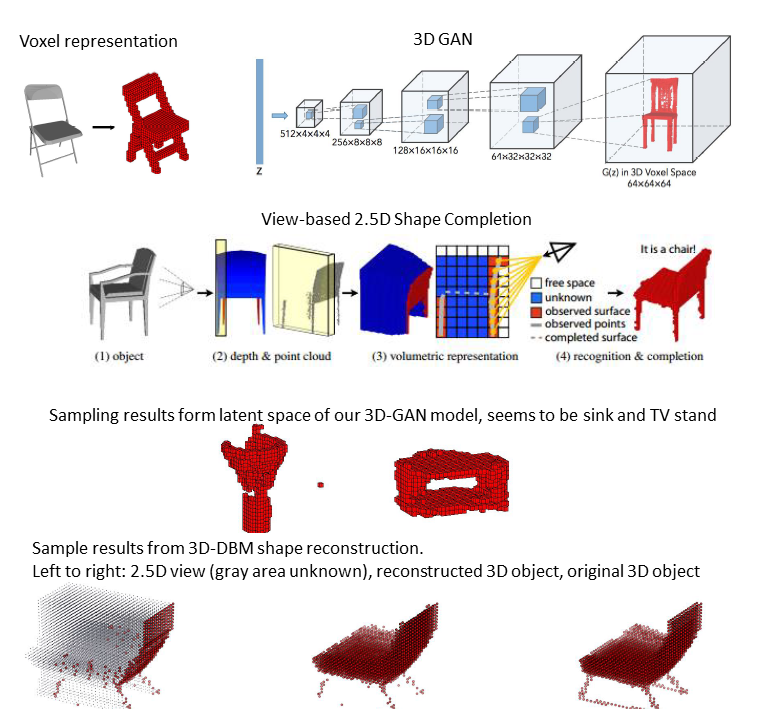
Deep Adversarial 3D Shape Net
Chieh Lin, Hsueh-Ti (Derek) Liu, Alex Kelly. Deep Adversarial 3D Shape Net [report]
This is the final project for course 10807-Topics in Deep Learning taken in 2016 Fall. In this project we have reviewed various deep learning models applied to 3D shape data. It has been shown that voxel representation is suitable for training deep learning models. The learned shape representation is able to perform classification and reconstruction given 2D or 2.5D images. However, how to ensure generated shapes are valid is still an unsolved problem in this field. We implemented 3D-CNN for classification, 3D-VAE and 3D-GAN for shape generation, and 3DDBM for reconstructing 3D shapes from 2.5D data. Our ultimate goal is to combine these approaches to build a 3D-AAE and modify the loss function to add reqularized term in order to generate more smooth and plausible shapes. Unfortunately, we could not obtain satisfactory results by the deadline. We failed to capture surface detail of the shapes, but we still achieve promising results in all three tasks we mentioned above.
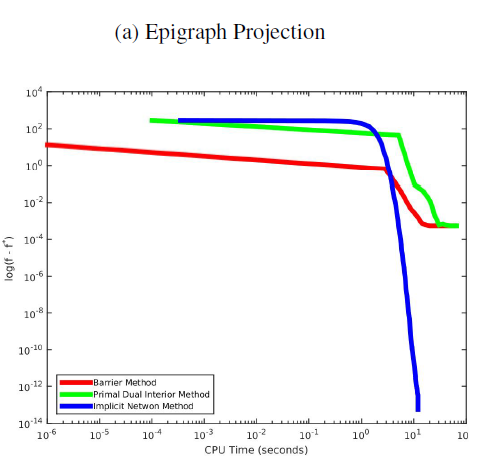
The Implicit Dual Newton Method
Po-Wei Wang, Chieh Lin, Tzu-Ming Kuo. The Implicit Dual Newton Method [report]
This is the final project for course 10725-Convex Optimization taken in 2016 Fall. To solve convex optimization problem, we can apply the primal-dual interior point method, which approximates the original objective f(x) and contraints gi(x) by second-order Taylor expansions, examines the KKT condition of the approximation, and then solves the KKT system by Newton method. On the other hand, we know that a dual problem corresponding to the prime problem exist. Assuming zero duality gap, can we simply solve the primal-dual pair without approximation? Unfortunately, unlike quadratic program or semi-definite program, there are no analytic dual problems for most convex primal problems. This makes the optimization involving the dual problem harder. However, we can show that, by implicit function theorem, there is a link between the primal and dual derivatives (Dontchev and Rockafellar, 2009). We can actually derive the gradient and Hessian for the dual problem from the proximal functions of f and gi, solving the dual optimization implicitly (Wang et al., 2016). For epigraph projection application, we can see that the implicit Newton method did converges faster than the other method.
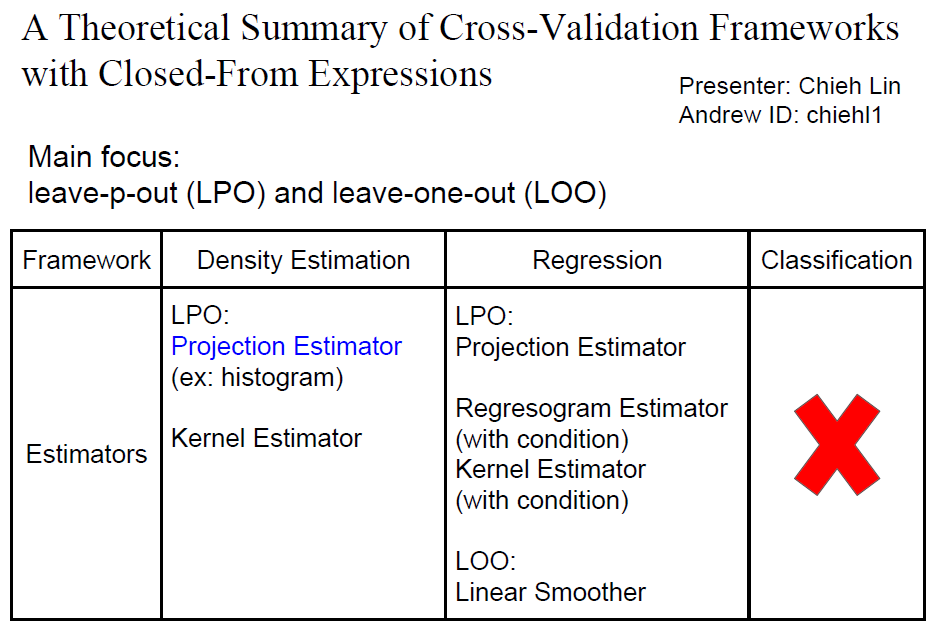
A Theoretical Summary of Cross-Validation Frameworks with Closed-From Expressions
Chieh Lin , A Theoretical Summary of Cross-Validation Frameworks with Closed-From Expressions [report, slides]
This is the final project for course 36702-Statistical Machine Learning taken in Spring 2016. For machine learning and statistical models, cross-validation (CV) method is widely used for model selection and performance estimation. The philosophy of CV is to split the limited amount of data into training set and validation set. The validation set can be the "new data" for estimating the performance of the model. Since CV only assumes that the data are identically distributed and the training set are independent from validation set, CV is universal and can be applied to almost all of frameworks such as regression, density estimation, and classification. The most popular CV schemes are leave-one-out (LOO) cross-validation, and K-fold cross-validation. However, even the most naive implementation of cross-validation with K data splits has a complexity of K times the model training time. This might be impossible to do depending on how expensive the model training is. This project aims to survey the frameworks that have closed-form cross-validation expressions and figure out the theoretical basis of each closed-form expression. To be more specific, we will focus on (LOO) and leave-p-out (LPO) cross-validation schemes where p can be any number between 1 and n-1 where n is the size of training set. With this information, the risks of the models built in these frameworks can be easily estimated by the studied closed-form expressions. From literature survey, it is known that closed-form formulas of LOO and LPO risk estimator have been derived for projection estimator, histogram estimator(which is a special case of projection estimator), and kernel estimators for density estimation. For least squares in linear regression, closed-form formulas for LPO estimators are also derived in regression for regressogram, kernel and projection estimators. For regressogram and kernel estimator in regression, additional assumption on data and the kernel functions need to be satisfied, which makes these two closed-forms not universal, so the detail of these two LPOs is not covered in this project. We have investigated and verified that closed-form LPO and LOO are available in density estimation and regression frameworks with some well-known estimators. This project provides a summary of the exact closed-form expressions under specific estimators and frameworks.
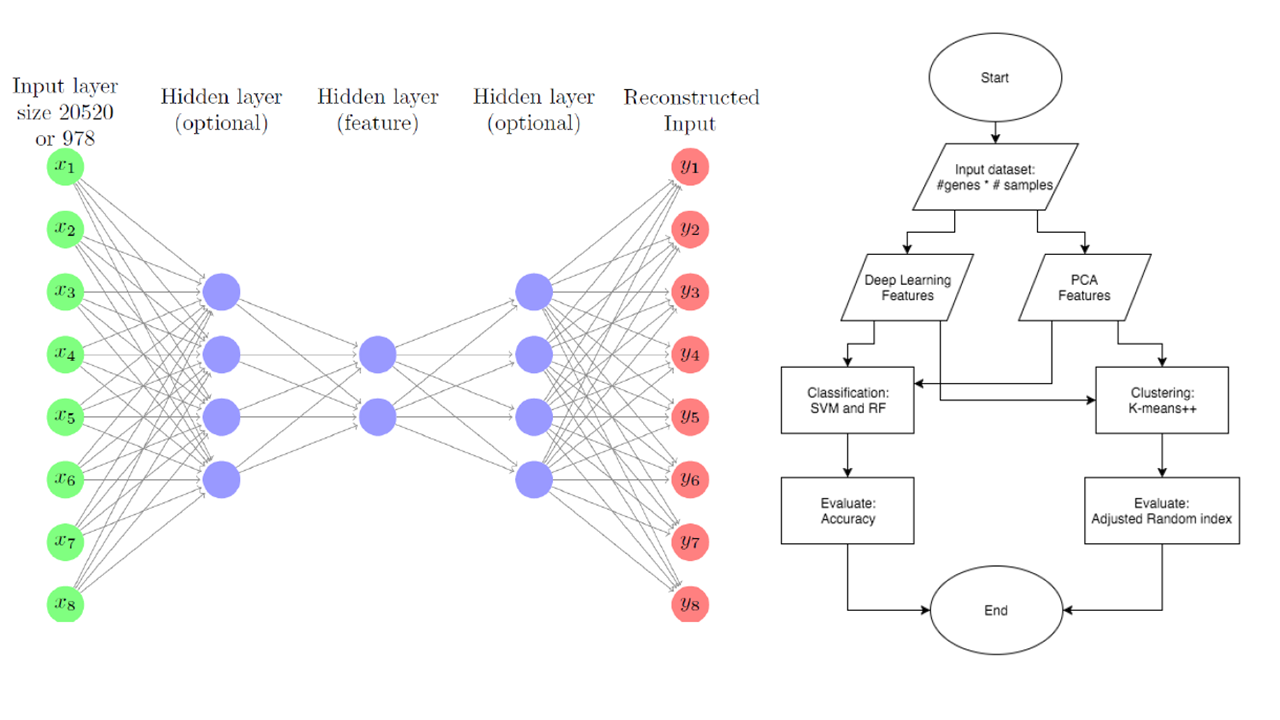
Classification and Clustering of Single Cell RNA-seq Gene Expression Data Using Deep Learning Features
Chieh Lin, Classification and Clustering of Single Cell RNA-seq Gene Expression Data Using Deep Learning Features [report, poster]
This is the final project for course 02710-Computational Genomics taken in Spring 2016. To understand the regulations of cell functions in complex biological systems, it is very important to identify various cell types in the systems. Take nervous systems as example, the cell type diversity is still poorly understood so that the most of the brain functions remain a mystery. Also, even the cells with the same cell type might play different roles in different phases. Therefore, classification of different phases of cells is also important. For now, several of the cell type classification works are based on principal component analysis (PCA) with manual curations. This project aims to apply deep learning methods to extract non-linear features from the single cell RNA-seq data and use the feature in other classification or clustering methods. The ultimate goal of this project is using deep learning to generate deep features (embeddings) to help classify/cluster unknown cell types in order to discover new cell types. From the results, it can be observed that PCA is slightly better than NN in classification but NN is much better than PCA in clustering. Since our ultimate goal is discovering new cell types, the result of clustering is more important than classification. We assume that NN can somehow learn non-linear biological information from the input and help clustering performance. Also, the expected result of landmark genes is no better than the set of all genes, and the result corresponds to our assumption. This result shows that if we use only landmark genes to speed up the model training time, we could lose important biological information that can help distinguish new cell types.
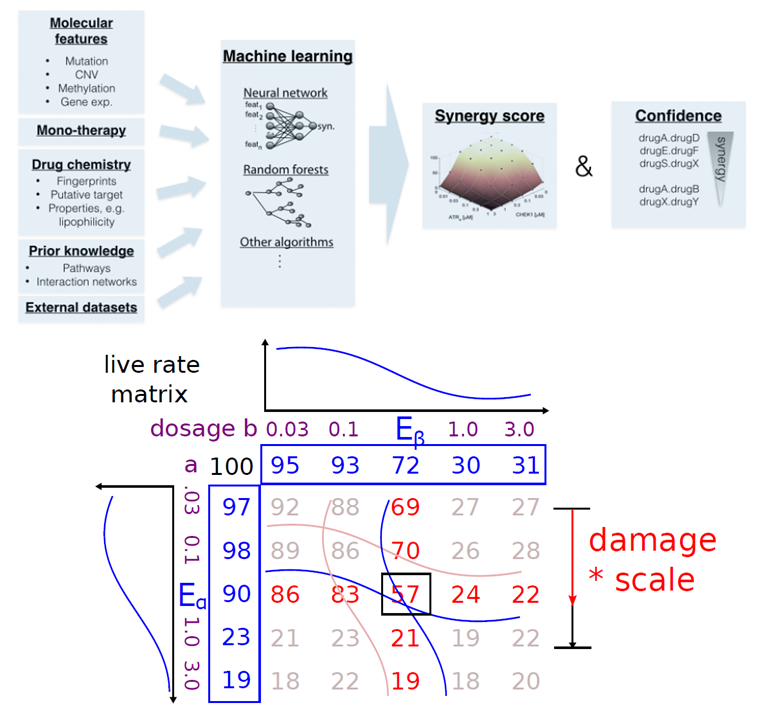
Drug Combination Prediction Challenge
Chieh Lin, Po-Wei Wang, Yan Xia (Not in the class), Ziv Bar-Joseph (Supervisor), Drug Combination Prediction Challenge [report, slides]
This is the final project for course 10715-Advanced Introduction to Machine Learning taken in Fall 2015. The multi-drug combination effects is always an important issue because it can help develop new therapies for diseases and prevent deadly drug-combination side-effects. However, it takes substantial efforts and resources for in vivo methods to verify the effects since the number of combinations is too large. Therefore, the in vitro method of machine learning for predicting the multi-drug combination effect is necessary. The AstraZeneca-Sanger Drug Combination Prediction Challenge [1] provides datasets for modelling two-drug synergy effect. With the datasets, the goal of this project is to find the best machine learning strategy to solve the two-drug synergy prediction problem, hoping to lay the foundation of future multi-drug combination effect prediction. For now, we have tried three strategies to deal with drug synergy problem: curve fitting, naive regression, and our newly developed locality model. We found that assuming the dataset is a sigmoidal curve and doing curve fitting results in the lowest performance. This tells us that although the sigmoidal curve may best characterize the behavior of dosage-response curve theoretically, the noises accumulated in each steps of experiment might still have a great impact to the performance. Logistic regression gives better results, but still no better than locality model. The locality model is the new way to predict live rate matrix from mono therapy without fitting any parametric curve. We developed new algorithm of proximal coordinate descent to solve the optimization problem of locality model with complexity linear to the number of features. Also, the L1 regulation term in regression plus poly2 expansion leads to good result with controllable non-zero terms, which shows that this technique can help keep the model from overfiting with stable performance in cross validation. This study is a systematic way to model drug synergy effect without using too much domain knowledge.
Computer-Aided Drug Design Project
This is the Final Project for the course Computer-Aided Drug Design taken in 2013. This project required us to choose a paper to implement. We implemented this paper "Physically Motivated, Robust, ab Initio Force Fields for CO2 and N2" with my team member Yu-Chuan Su. Specifically, we perform molecular dynamic (MD) simulations with small molecules of CO2 and N2 with various force fields.
Chinese Dark Chess A.I.
This is the Final Project for the course Theory of Computer Games taken in 2011. I use negascout algorithm, transposition table, move ordering, scoring function, and assigning different pieces with different value to achieve this A.I. The red player is A.I., and black player is me. The video shows part of my game with my A.I.
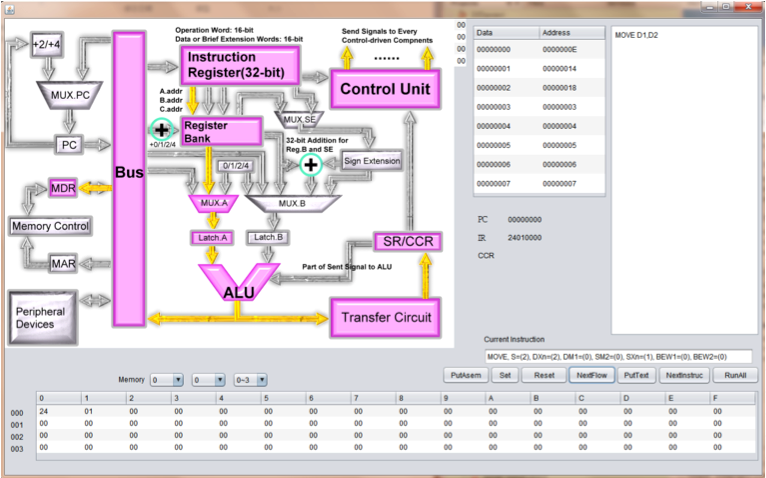
M68000 CPU instruction set implementation
This is the Final Project for the course Digital Circuit Design taken in 2011. In this project, we are asked to implement about 50 instructions from M68000 CPU. We choose about 50 instructions and 6 addressing mode of to implement (This is a total about 300 instructions). We design the block diagram for these instructions and write a simulator, assmbler with gui demonstrating the data flow for each instruction.
Intelligent Car
This is the Final Project for the course Digital Circuit Laboratory taken in 2011. We use DE2 FPGA board as the core of our car and output signals to GWS-S35 servo mortors to drive the car. The car will do corresponding actions when it sees specific colors. Blue: moving forward, Yellow: moving backward, Green: turning left/right
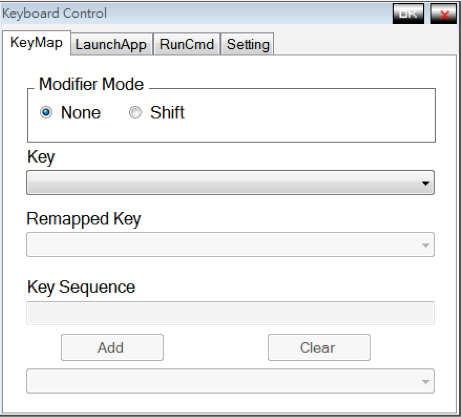
Keyboard Agent
This is a Project when I was work as a programmer in Nexcom, 2010. This program let users define their key mapping preference. The pre-defined keys can be mapped to a sequence of keys, launching applications and running commands.
Game Projects
We use Java and C to implement games in Object-Oriented Programming course and Assembly course taken in 2010 as my final project. The video is a single-player version Puyo Puyo game.